This function generates plots to visualize model performance
Usage
performance_plot(
model_list,
type = "box",
text_size = 10,
palette = "viridis",
save = FALSE,
file_path = NULL,
file_name = "Performance_plot",
file_type = "pdf",
plot_width = 7,
plot_height = 7,
dpi = 80
)Arguments
- model_list
A
model_listobject from performingtrain_models.- type
Type of plot to generate. Choices are "box" or "dot." Default is
"box."for boxplots.- text_size
Text size for plot labels, axis labels etc. Default is
10.- palette
Viridis color palette option for plots. Default is
"viridis". Seeviridisfor available options.- save
Logical. If
TRUEsaves a copy of the plot in the directory provided infile_path.- file_path
A string containing the directory path to save the file.
- file_name
File name to save the plot. Default is
"Performance_plot."- file_type
File type to save the plot. Default is
"pdf".- plot_width
Width of the plot. Default is
7.- plot_height
Height of the plot. Default is
7.- dpi
Plot resolution. Default is
80.
Details
The default metrics used for classification based models are "Accuracy" and "Kappa."
These metric types can be changed by providing additional arguments to the
train_modelsfunction. SeetrainandtrainControlfor more information.
Examples
# \donttest{
## Create a model_df object
covid_model_df <- pre_process(covid_fit_df, covid_norm_df)
#> Total number of differentially expressed proteins (8) is less than n_top.
#> None of the proteins show high pair-wise correlation.
#>
#> No highly correlated proteins to be removed.
## Split the data frame into training and test data sets
covid_split_df <- split_data(covid_model_df)
## Fit models based on the default list of machine learning (ML) algorithms
covid_model_list <- train_models(covid_split_df)
#>
#> Running svmRadial...
#> Loading required package: ggplot2
#> Loading required package: lattice
#>
#> Running rf...
#>
#> Running glm...
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: algorithm did not converge
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#>
#> Running xgbLinear...
#>
#> Running naive_bayes...
#> Done!
## Generate box plots to visualize performance of different ML algorithms
performance_plot(covid_model_list)
#> Using Resample as id variables
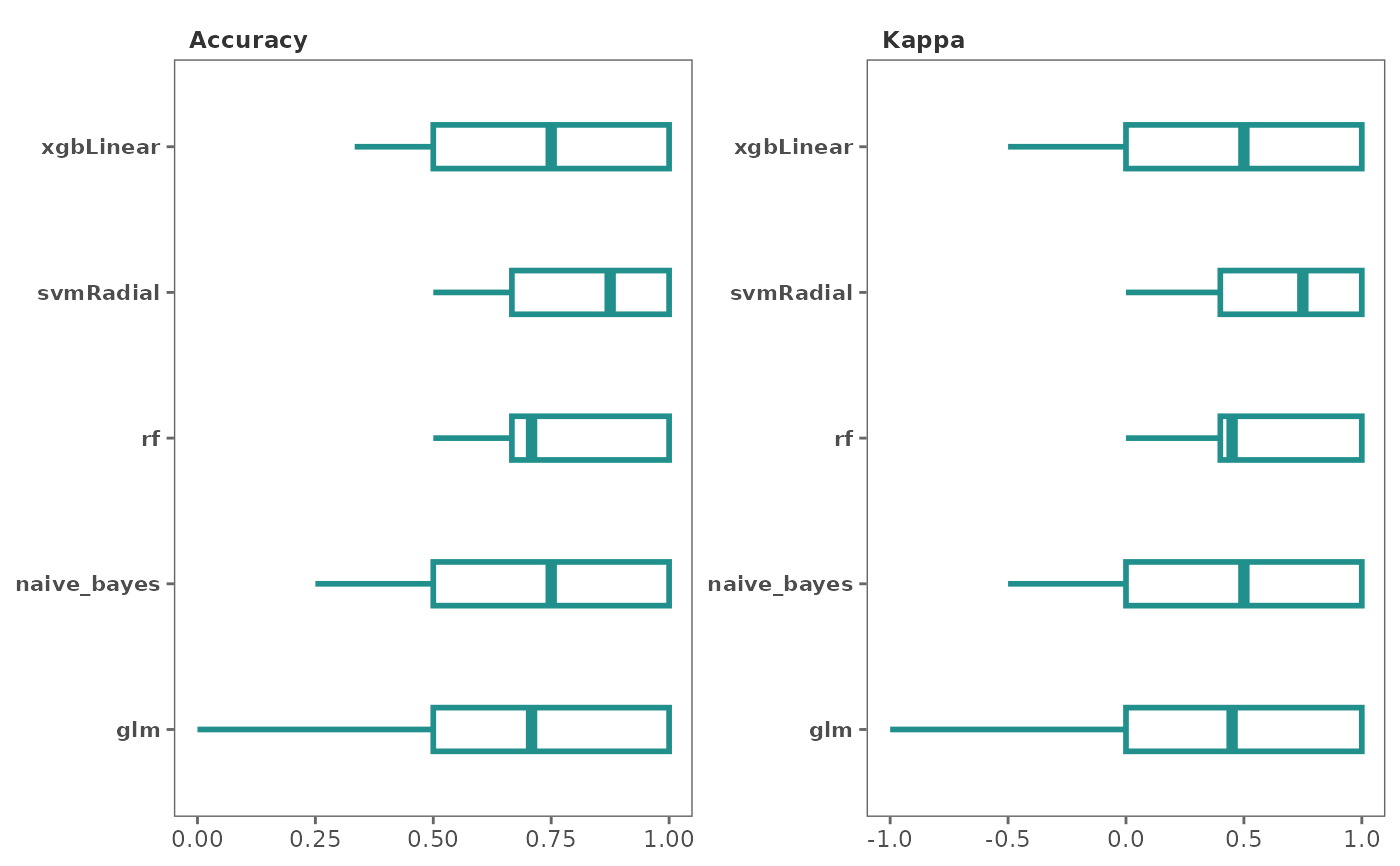 ## Generate dot plots
performance_plot(covid_model_list, type = "dot")
#> Using Resample as id variables
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
## Generate dot plots
performance_plot(covid_model_list, type = "dot")
#> Using Resample as id variables
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
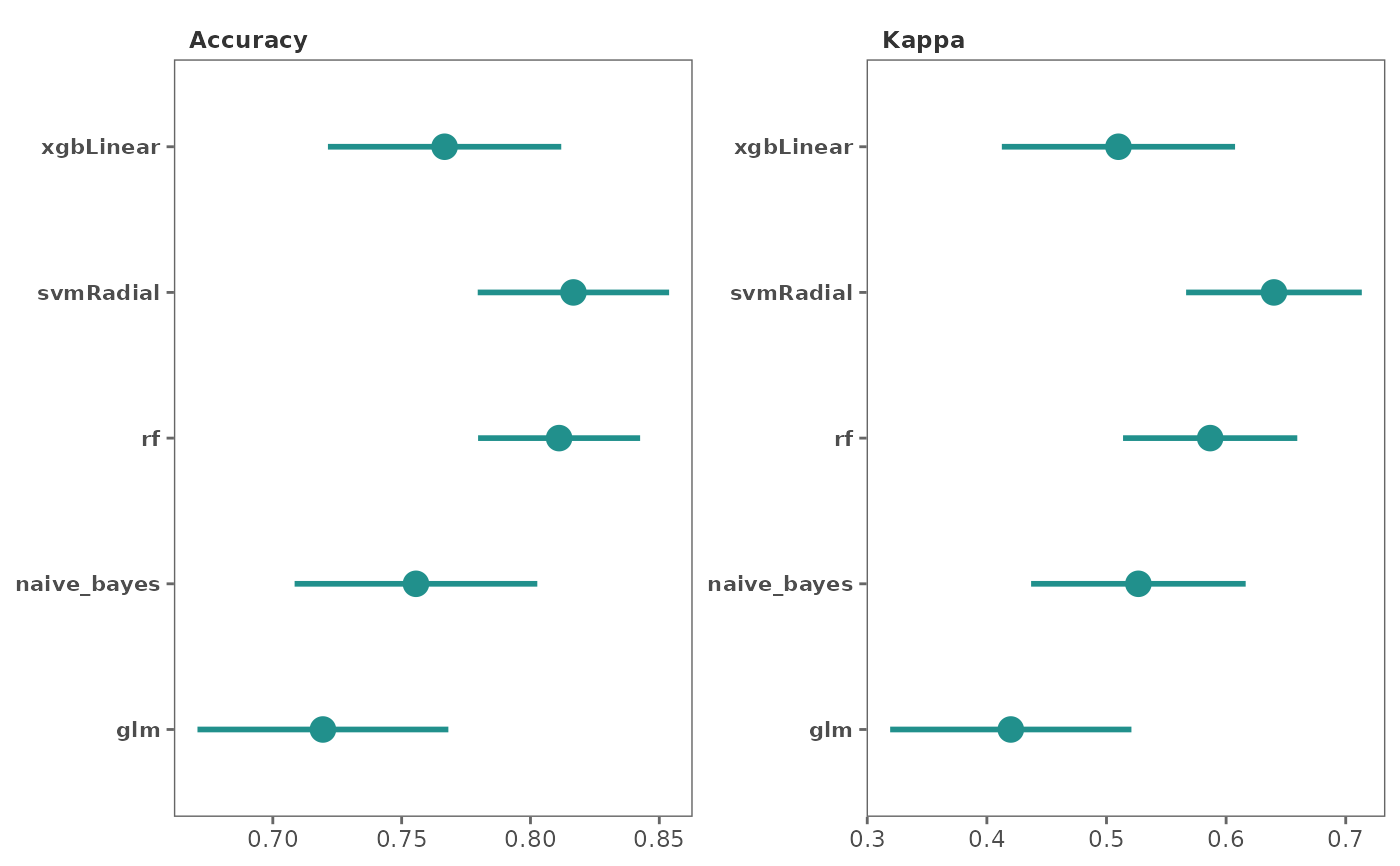 ## Change color palette
performance_plot(covid_model_list, type = "dot", palette = "inferno")
#> Using Resample as id variables
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
## Change color palette
performance_plot(covid_model_list, type = "dot", palette = "inferno")
#> Using Resample as id variables
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
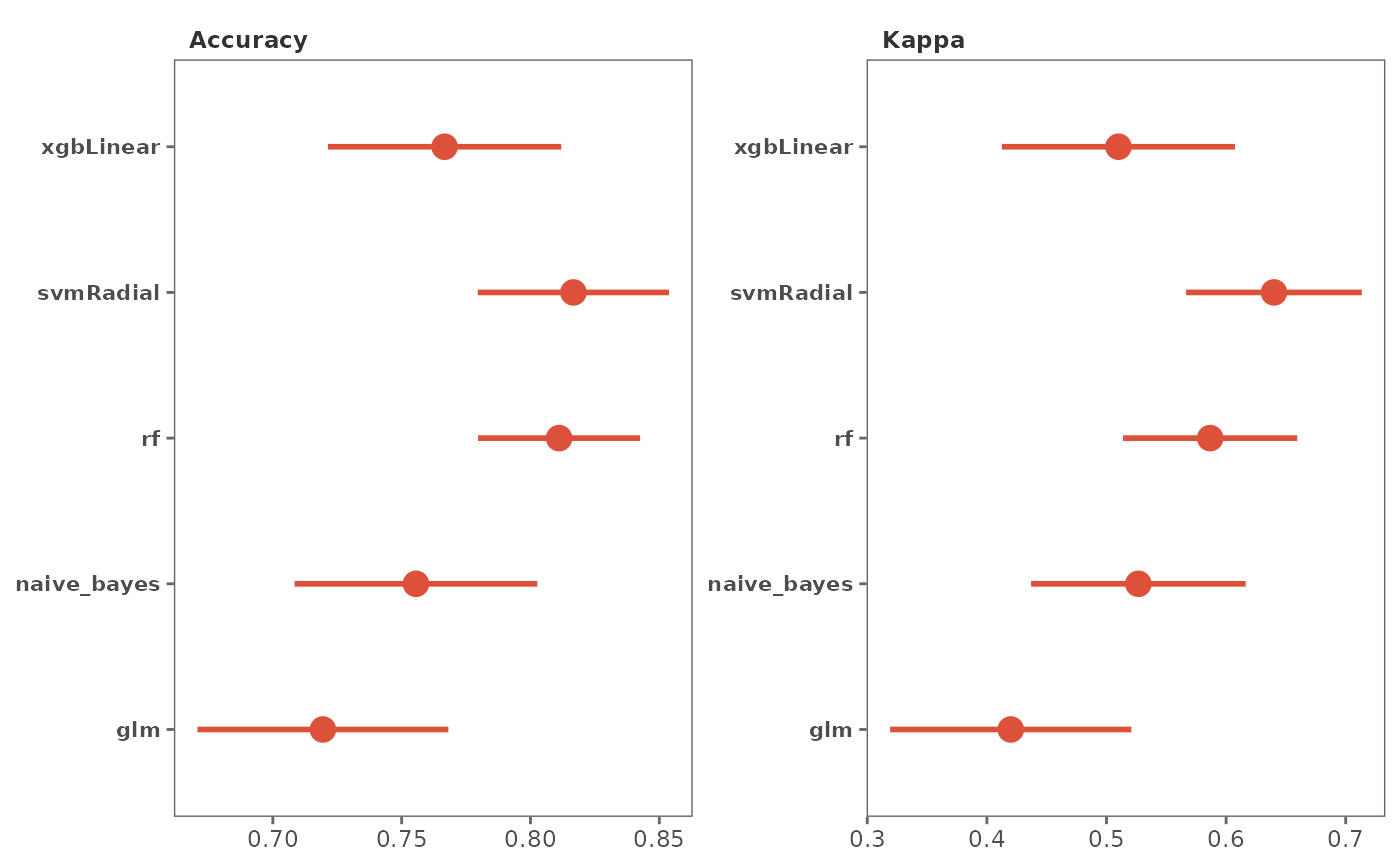 # }
# }
