promor
An R package for proteomics data analysis and machine learning based predictive modeling

This project was perhaps my most ambitious one to date. When I first started working as a Computational Biologist at the Eastern Virginia Medical School, I had just made the transition from doing work that involved both wet lab and computational work to purely computational biology/ bioinformatics. I, along with two other colleagues, were hired to work primarily on a project called the Digital Patient at EVMS. Within the overarching goals of that project, we had the opportunity to design our own projects that would be of interest to the faculty at the school and would benefit their ongoing research. While working with a research group exploring proteomics-based biomarkers for early detection of prostate cancer, I thought of developing a custom bioinformatics pipeline to cater the group’s biomarker research. promor was built based on those initial efforts. With no experience whatsoever in software development of any kind, let alone R package development, I had a lot to learn about the best practices in the process. Looking back, I am surprised at how quickly the package came together - within just a few short months, promor made it to CRAN and a publication was in the works. Now, I am excited to see the package being used by research groups from around the world, and recently, promor passed 7000 downloads on CRAN.
Motivation
I was motivated to bulid promor to streamline the transition from identifying differentially expressed proteins in proteomics data to using them in predictive models for disease diagnosis or prognosis. In many proteomics studies, it is not unusual to identify dozens or even hundreds of differentially expressed proteins between groups of interest (e.g., cancer stages or cancer vs. non-cancer patients). However, including all of them in a diagnostic test is often impractical and not useful. The challenge lies in selecting robust candidates that effectively stratify patient populations, which has led many scientists to turn to machine learning-based models. Existing proteomics data analysis tools lacked an efficient and reproducible workflow to streamline this process, and promor was designed to fill that gap (Ranathunge et al., 2023).
Proteomics data analysis
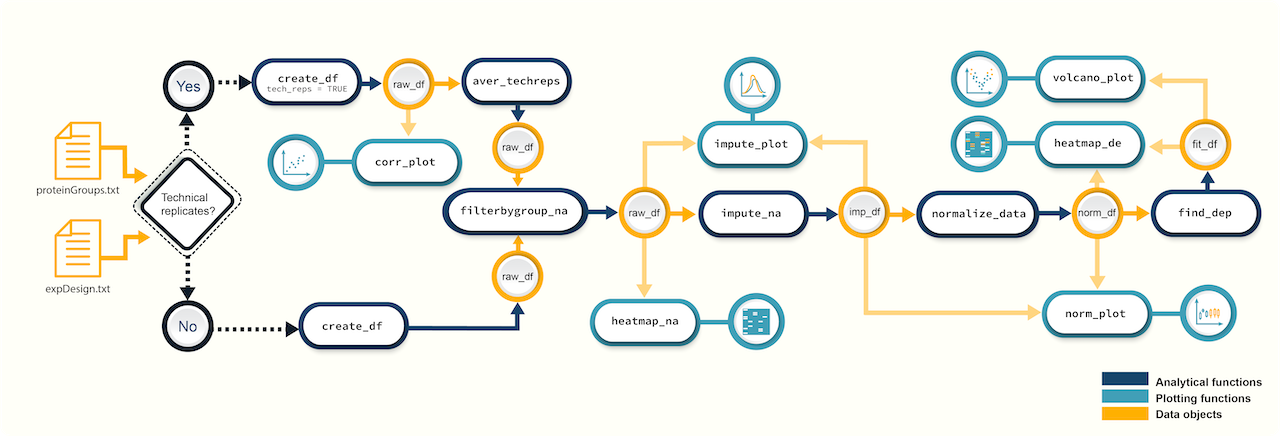
promor’s workflow for proteomics data analysis includes functions for quality control, visualization, and differential expression analysis. It also has the capability to handle technical replicates in the data. Visit the promor websitefor more information and step-by-step tutorials.
Predictive modeling
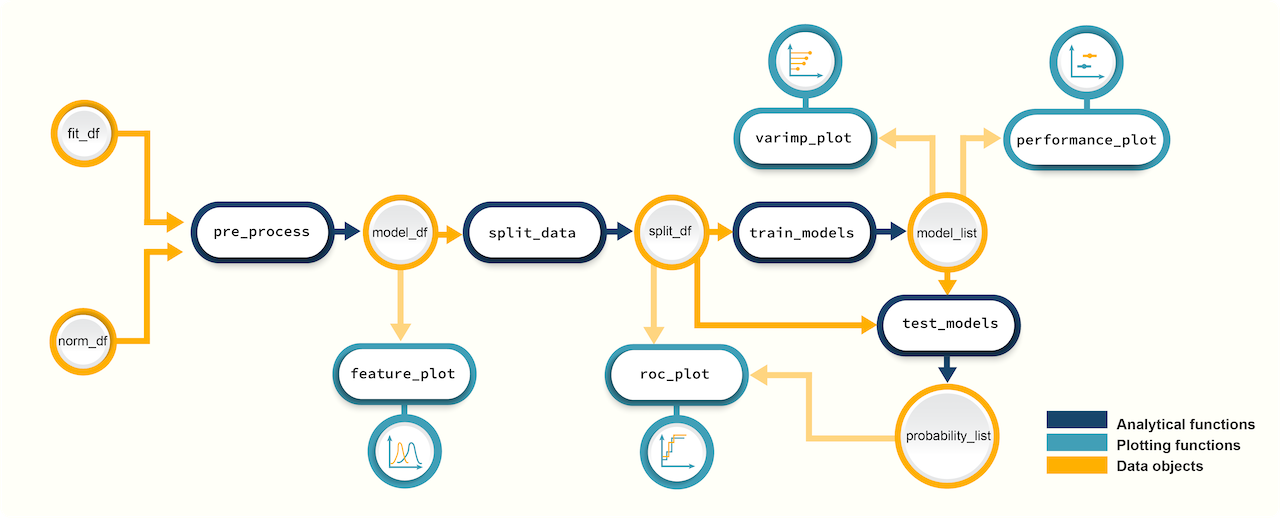
promor’s workflow for building machine learning based predictive models provides access to over 200 ML algorithms via the R package caret used in the backend. In addition to model building, this workflow includes functions for quality control and visualization (e.g. feature plots, variable importance plots, ROC curves). For more information and detailed tutorials, visit the promor website.

