Short Tandem Repeats
The functional and adaptive role of short tandem repeats

My dissertation focused on the functional and adaptive role of microsatellites, and to investigate their role I used natural populations of common sunflower as a model system. Microsatellites, also called short tandem repeats (STRs), are highly mutable and abundant throughout eukaryotic genomes. While their role in human diseases is well-documented, their potential adaptive role is still debated. One way microsatellites may contribute to adaptation is through their influence on gene expression. We tested this hypothesis on natural populations of the common sunflower (Helianthus annuus), a species that is widespread across much of North America and well adapted to its local environments.
Transcribed microsatellites (STRs) linked to gene expression variation in sunflowers
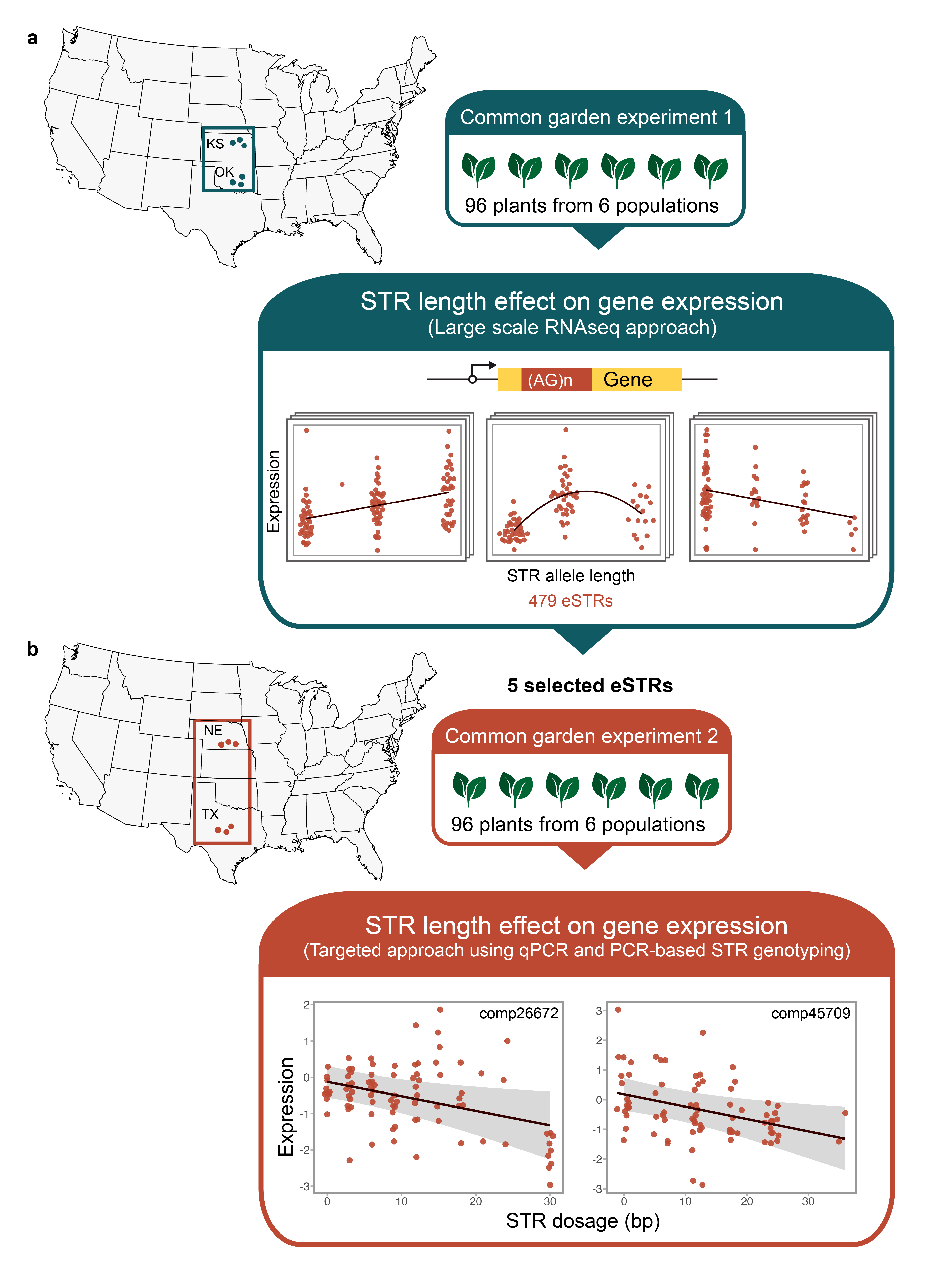
Through our work, we found substantial evidence that transcribed microsatellites (eSTRs) can impact gene expression levels. For example, our study using RNA-seq data from 95 individuals from different sunflower populations identified 479 eSTRs where allele length was significantly correlated with gene expression . Notably, these correlations often exhibited a stepwise pattern, supporting the “tuning knob model”, which proposes that gradual changes in microsatellite length can fine-tune gene expression and ultimately influence phenotypic traits. Our results also showed that most of the eSTRs were located in the UTRs, in locations ideal for regulating gene expression (Ranathunge et al., 2020).
eSTRs are under purifying selection
We also compared this special group of eSTRs to anonymous STRs within the sunflower, looking for signatures of selection, which would solidify our hypothesis that these eSTRs are indeed important in sunflower adaptation. We found that compared to anonymouss or predominantly neutral STRs, these eSTRs showed lower allelic diversity, heterozygozity and allelic richness, indicating higher relative rates of purifying selection. We also found that at some eSTRs, allele length significantly correlated with latitude, which aligned with the predictions of the tuninig knob hypthesis (Ranathunge et al., 2022).
The role of microsatellites (STRs) in population and species divergence
We also explored the role of microsatellites in gene expression divergence among closely related Helianthus species. We compared transcriptomes of 50 individuals from five species and discovered a significant pattern: genes containing microsatellites in non-coding regions, such as UTRs, were more likely to be differentially expressed between species than genes with microsatellites in coding regions or those lacking microsatellites. This trend was consistent across all species pairs, suggesting non-coding microsatellites may be crucial in shaping gene expression divergence during speciation. We also observed greater genetic divergence in transcripts containing microsatellites compared to those without, as measured by pairwise FST at SNPs. This suggests that genes with microsatellites may be under different selective pressures, leading to increased genetic differentiation among species (Ranathunge et al., 2021).
Further evidence points to the influence of microsatellites on gene expression divergence among populations of the same species. We found that microsatellite-containing differentially expressed genes were related to transcription regulation and transcription factor activity and that genes containing repeats of A or AG are more likely to be differentially expressed among populations from a latitudinal cline (Ranathunge et al., 2018). This suggests microsatellites may play a key role in regulating genes involved in important biological processes that contribute to population and species differentiation.


Clinal patterns of variation in eSTRs in sunflower
To further investigate the relationship between microsatellite length variation and gene expression, we conducted a targeted study focusing on five eSTRs in Helianthus annuus populations across a latitudinal gradient. We found a significant correlation between eSTR length and gene expression for an eSTR located within the CHUP1 gene, which is involved in chloroplast movement (Ranathunge & Welch, 2024). This finding suggests that this specific eSTR might contribute to sunflower adaptation across different environments and that longer or shorter alleles may be favored in extreme environmental conditions,
Our research, combined with that of others, strongly suggests that STRs, particularly those located in transcribed regions, can contribute to both gene expression variation within populations and gene expression divergence between species. Our work highlights the potential importance of these often-overlooked genomic elements in adaptation and evolution.









